Welcome to Zacharias Web Services!
The Public Web Server of the T38 Workgroup
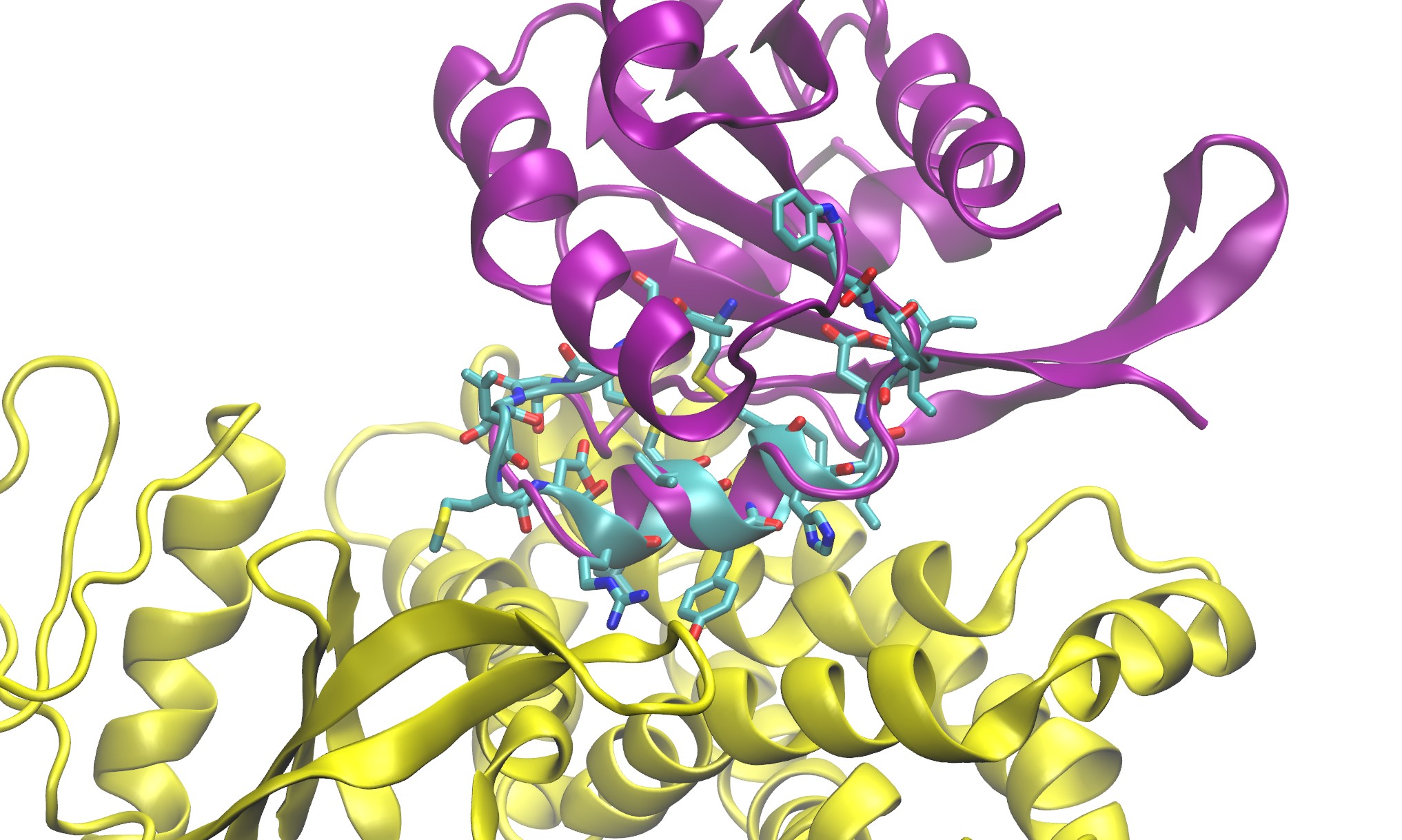
The function of proteins and nucleic acids in living systems is strongly coupled to the molecular motion and dynamics of these biomolecules. We use computer simulation methods to study the structure, function and dynamics of biomolecules. Our goal is to better understand structure formation processes and to elucidate the mechanism of ligand-receptor association in atomic detail.
We offer some of our simulations here for public use. Please look through our simulation catalog and see if you can find the right service for you.
We run your files on our cluster and return you your final product via email.
Please, check our User Guide in order to become familiar with the submission service.
References
Please cite the following reference if you use our simulations:
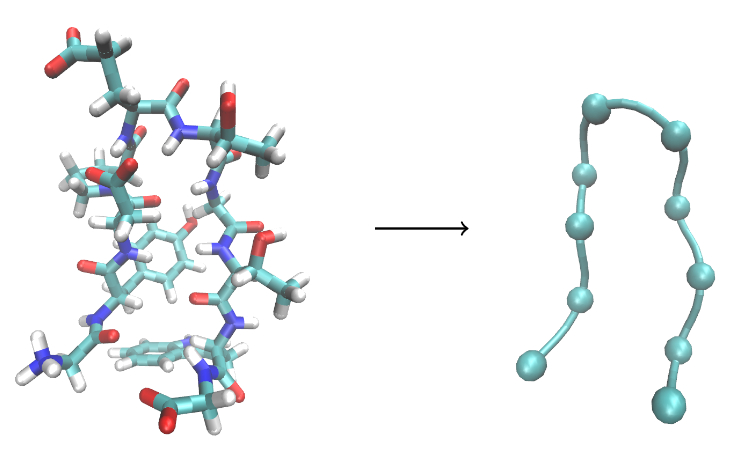
Cyclic Peptide Matching Algorithm:
Santini BL and Zacharias M;
Rapid in silico Design of Potential Cyclic Peptide Binders Targeting Protein-Protein Interfaces.
Front. Chem. , 2020.
DOI: 10.3389/fchem.2020.573259
Replica Exchange with Repulsive Scaling:
Siebenmorgen T, Engelhard M and Zacharias M;
Prediction of protein–protein complexes using replica exchange with repulsive scaling.
J Comput Chem. 2020.
DOI: 10.1002/jcc.26187
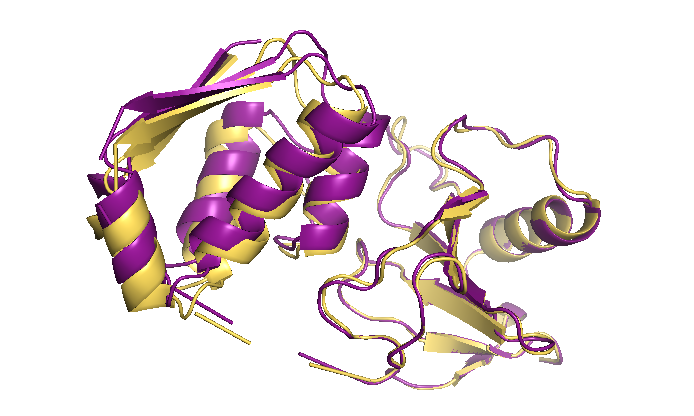
GoCa – Structure-based Coarse-grained Simulations:
Under Review